Why Glycans?
Glycosylation is the most common post-translational modification on proteins, and over half of all secreted proteins are expected to be glycosylated. Glycosylation may alter protein structure and dynamics, and thus affect enzyme activity, protein-protein interactions, and the in vivo circulation half-life of protein pharmaceuticals. In addition, glycans are also involved in cellular recognition by interacting with specific protein. At this time, however, it is difficult to understand which glycans are important components in the protein function and how to modify those glycans to optimize the protein properties of interest. To be able to predict a glycan’s impact on the protein function, it is critical to understand its impact on protein structure and dynamics.
Statistics of PDB glycans
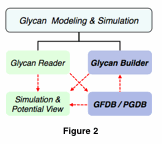
As of January 2011, there are 4,029 PDB structures (6.0%) with at least one glycan chain, yielding a total of 15,669 glycan chains (Figure 1, left). 8,848 of them (56%) are N-glycosylated, 688 (4.3%) are O-glycosylated, and the rest (6,133 chains, 39%) exists as ligands. The number of PDB structures with glycans deposited each year into the PDB shows steady increase over time (Figure 1, middle). However, the number of glycan chains as a function of glycan chain length (i.e., number of monosaccharide residues in a chain) shows that most glycan structures in PDB contain only one or two monosaccharides mainly due to removal of glycans prior to structural studies or due to crystallization conditions (Figure 1, right). These short glycan chains generally do not represent what is naturally found on these proteins. Fortunately, the “correct” glycan composition and linkage information is currently determinable by mass spectrometry. Therefore, a knowledge gap exists between knowing the glycan composition for a protein and understanding how that composition impacts the protein’s structure and dynamics. Taken together, these facts clearly support the critical needs for glycan modeling in structural glycobiology.
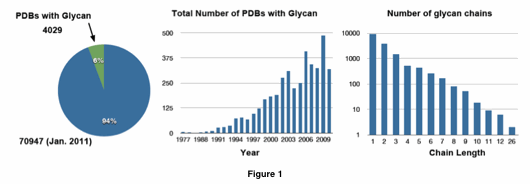
GlycanStructure portal for a unified computational facility for glycan modeling and simulation
An overview of this portal is shown in Figure 2. Glycan Reader is used to build Glycan Fragment DB and Glycan-Protein DB. Glycan Builder generates glycan structures through fragment-based threading approaches. This portal is tightly integrated with CHARMM-GUI for MD simulation input generation and online electrostatic potential visualization. Our innovative visions for glycan modeling and simulation will provide users with a one-stop-shop, where they can easily select glycan structures from the PDB, or construct their own glycans, and then use the CHARMM-GUI functional modules for simulation and electrostatic potential visualization.